[Publications]
[Publications]
(46)
Minagawa, Y., Hata, M., Yamamoto, E., Tsuzuki, D., Morimoto, S., (2023).
Inter-brain synchrony during mother-infant interactive parenting in 3-4-month-old infants with and without an elevated likelihood of autism spectrum disorder.
Cerebral Cortex, bhad395, https://doi.org/10.1093/cercor/bhad395
(45)
Inter-brain synchrony during mother-infant interactive parenting in 3-4-month-old infants with and without an elevated likelihood of autism spectrum disorder.
Cerebral Cortex, bhad395, https://doi.org/10.1093/cercor/bhad395
Su, W.-C., Culotta, M., Mueller, J., Tsuzuki, D., Bhat, A, (2023).
Autism-Related Differences in Cortical Activation When Observing, Producing, and Imitating Communicative Gestures: An fNIRS Study.
Brain Sciences., 13, no. 9: 1284. https://doi.org/10.3390/brainsci13091284
(44)
Autism-Related Differences in Cortical Activation When Observing, Producing, and Imitating Communicative Gestures: An fNIRS Study.
Brain Sciences., 13, no. 9: 1284. https://doi.org/10.3390/brainsci13091284
Su, W.-C., Culotta, M., Mueller, J., Tsuzuki, D., Bhat, A, (2023).
fNIRS-Based Differences in Cortical Activation during Tool Use, Pantomimed Actions, and Meaningless Actions between Children with and without Autism Spectrum Disorder (ASD).
Brain Sciences., 13, no. 6: 876. https://doi.org/10.3390/brainsci13060876
(43)
fNIRS-Based Differences in Cortical Activation during Tool Use, Pantomimed Actions, and Meaningless Actions between Children with and without Autism Spectrum Disorder (ASD).
Brain Sciences., 13, no. 6: 876. https://doi.org/10.3390/brainsci13060876
Tsuzuki, D.*, Taga, G., Watanabe, H. & Homae, F., (2022).
Individual variability in the nonlinear development of the corpus callosum during infancy and toddlerhood: a longitudinal MRI analysis.
Brain Structure and Function., doi:10.1007/s00429-022-02485-y
(42)
Individual variability in the nonlinear development of the corpus callosum during infancy and toddlerhood: a longitudinal MRI analysis.
Brain Structure and Function., doi:10.1007/s00429-022-02485-y
Su, W.C., Culotta, M., Tsuzuki, D. & Bhat, A.N., (2022).
Cortical activation during cooperative joint actions and competition in children with and without an autism spectrum condition (ASC): an fNIRS study
Sci Rep., 2022;12(1):5177. Published 2022 Mar 25. doi:10.1038/s41598-022-08689-w
(41)
Cortical activation during cooperative joint actions and competition in children with and without an autism spectrum condition (ASC): an fNIRS study
Sci Rep., 2022;12(1):5177. Published 2022 Mar 25. doi:10.1038/s41598-022-08689-w
Su, W.C., Culotta, M., Tsuzuki, D. & Bhat, A. N., (2021).
Movement kinematics and cortical activation in children with and without autism spectrum disorder during sway synchrony tasks: an fNIRS studySci Rep., 2021;11(1):15035. Published 2021 Jul 22. doi:10.1038/s41598-021-94519-4
(40)
Movement kinematics and cortical activation in children with and without autism spectrum disorder during sway synchrony tasks: an fNIRS studySci Rep., 2021;11(1):15035. Published 2021 Jul 22. doi:10.1038/s41598-021-94519-4
Su, W. C., Culotta, M., Mueller, J., Tsuzuki, D., Pelphrey, K. & Bhat A. N., (2020).
Differences in cortical activation patterns during action observation, action execution, and interpersonal synchrony between children with or without autism spectrum disorder (ASD): An fNIRS pilot study
PLoS One., 2020;15(10):e0240301. Published 2020 Oct 29. doi:10.1371/journal.pone.0240301
(39)
Differences in cortical activation patterns during action observation, action execution, and interpersonal synchrony between children with or without autism spectrum disorder (ASD): An fNIRS pilot study
PLoS One., 2020;15(10):e0240301. Published 2020 Oct 29. doi:10.1371/journal.pone.0240301
Su, W. C., Culotta, M. L., Hoffman, M. D., Trost, S. L., Pelphrey, K. A., Tsuzuki, D. & Bhat A. N., (2020).
Developmental Differences in Cortical Activation During Action Observation, Action Execution and Interpersonal Synchrony: An fNIRS Study
Frontiers in Human Neuroscience, 14, 57
(38)
Developmental Differences in Cortical Activation During Action Observation, Action Execution and Interpersonal Synchrony: An fNIRS Study
Frontiers in Human Neuroscience, 14, 57
Uchida, M., Arimitsu, T., Tsuzuki, D., Dan, I., Ikeda, K., Takahashi, T. & Minagawa, Y., (2019).
Maternal speech shapes the cerebral frontotemporal network in neonates: a hemodynamic functional connectivity study
Developmental Cognitive Neuroscience, 100701
(37)
Maternal speech shapes the cerebral frontotemporal network in neonates: a hemodynamic functional connectivity study
Developmental Cognitive Neuroscience, 100701
Sugiura, L., Hata, M., Matsuba-Kurita, H., Uga, M., Tsuzuki, D., Dan, I., Hagiwara, H. & Homae F., (2018).
Explicit Performance in Girls and Implicit Processing in Boys: A Simultaneous fNIRS-ERP Study on Second Language Syntactic Learning in Young Adolescents
Frontiers in Human Neuroscience, 12, 62
(36)
Explicit Performance in Girls and Implicit Processing in Boys: A Simultaneous fNIRS-ERP Study on Second Language Syntactic Learning in Young Adolescents
Frontiers in Human Neuroscience, 12, 62
Bhat, A., Hoffman, M., Trost, S., Culotta, M., Eilbott, J., Tsuzuki, D. & Pelphrey, K., (2017).
Cortical Activation in Adults during Action Observation, Action Execution, & Interpersonal Synchrony: A functional Near-Infrared Spectroscopy (fNIRS) Study
Frontiers in Human Neuroscience, 11, 431
(35)
Cortical Activation in Adults during Action Observation, Action Execution, & Interpersonal Synchrony: A functional Near-Infrared Spectroscopy (fNIRS) Study
Frontiers in Human Neuroscience, 11, 431
Tsuzuki, D., Homae, F., Taga, G., Watanabe, H., Matsui, M. & Dan, I., (2017).
Macroanatomical landmarks featuring junctions of major sulci and fissures and scalp landmarks based on the international 10-10 system for analyzing lateral cortical development of infants
Frontiers In Neuroscience, 11, 394.
(34)
Macroanatomical landmarks featuring junctions of major sulci and fissures and scalp landmarks based on the international 10-10 system for analyzing lateral cortical development of infants
Frontiers In Neuroscience, 11, 394.
Tsuzuki, D.*, Watanabe, H., Dan, I. & Taga, G., (2016).
MinR 10/20 system: Quantitative and reproducible cranial landmark setting method for MRI based on minimum initial reference points
Journal of Neuroscience Methods, 264, 86-93
(33)
MinR 10/20 system: Quantitative and reproducible cranial landmark setting method for MRI based on minimum initial reference points
Journal of Neuroscience Methods, 264, 86-93
Sugiura, L., Ojima, S., Matsuba-Kurita, H., Dan, I., Tsuzuki, D., Katura, T. & Hagiwara, H., (2015).
Effects of sex and proficiency in second language processing as revealed by a large-scale fNIRS study of school-aged children
Human Brain Mapping, 36(10), 3890-3911
(32)
Effects of sex and proficiency in second language processing as revealed by a large-scale fNIRS study of school-aged children
Human Brain Mapping, 36(10), 3890-3911
Monden, A., Dan, I., Nagashima, M., Dan, H., Uga, M., Ikeda, T., Tsuzuki, D., Kyutoku Y., Gunji, Y., Hirano, D., Taniguchi, T., Shimoizumi, H., Watanabe, E. & Yamagata, T., (2015).
Individual classification of ADHD children by right prefrontal hemodynamic responses during a go/no-go task as assessed by fNIRS
NeuroImage: CLINICAL, 9, 1-12
(31)
Individual classification of ADHD children by right prefrontal hemodynamic responses during a go/no-go task as assessed by fNIRS
NeuroImage: CLINICAL, 9, 1-12
Rizki, E., Uga, M., Dan, I., Dan, H., Tsuzuki, D., Mizutani, T., Sano, T., Yokota, H., Oguro, K., Watanabe, E., (2015).
Determination of epileptic focus side in mesial temporal lobe epilepsy using long-term non-invasive fNIRS/EEG monitoring for presurgical evaluation
Neurophotonics, 2(2). 025003
(30)
Determination of epileptic focus side in mesial temporal lobe epilepsy using long-term non-invasive fNIRS/EEG monitoring for presurgical evaluation
Neurophotonics, 2(2). 025003
Aasted, CM., Yücel, MA., Cooper, RJ., Dubb, J., Tsuzuki, D., Becerra, L, Petkov, MP., Borsook, D., Dan, I., Boas, DA., (2015).
Anatomical Guidance for Functional Near-Infrared Spectroscopy: An AtlasViewer Tutorial.
Neurophotonics, 2(2). 020801
(29)
Anatomical Guidance for Functional Near-Infrared Spectroscopy: An AtlasViewer Tutorial.
Neurophotonics, 2(2). 020801
Nagashima, M., Monden, Y., Dan, I., Dan, H., Mizutani, T., Tsuzuki, D., Kyutoku, Y., Gunji, Y., Hirano, D., Taniguchi, T., Shimoizumi, H., Momoi, M.Y., Yamagata, T., & Watanabe, E., (2014).
Neuropharmacological effect of atomoxetine on attention network in children with attention deficit hyperactivity disorder during oddball paradigms as assessed using functional near-infrared spectroscopy
Neurophotonics, 1(2). 025007
(28)
Neuropharmacological effect of atomoxetine on attention network in children with attention deficit hyperactivity disorder during oddball paradigms as assessed using functional near-infrared spectroscopy
Neurophotonics, 1(2). 025007
Nagashima, M., Monden, Y., Dan, I., Dan, H., Tsuzuki, D., Mizutani, T., Kyutoku, Y., Gunji, Y., Hirano, D., Taniguchi, T., Shimoizumi, H., Watanabe, E., Momoi, M.Y., & Yamagata, T, (2014).
Acute neuropharmacological effects of atomoxetine on inhibitory control in ADHD children: A fNIRS study
NeuroImage: CLINICAL, 6. 192-201
(27)
Acute neuropharmacological effects of atomoxetine on inhibitory control in ADHD children: A fNIRS study
NeuroImage: CLINICAL, 6. 192-201
Nagashima, M., Dan, I., Monden, Y., Dan, H., Tsuzuki, D., Mizutani, T., Kyutoku, Y., Gunji, Y., Momoi, Y. M., Watanabe, E. & Yamagata, T., (2014).
Neuropharmacological effect of methylphenidate on attention network in children with attention deficit/hyperactivity disorder during oddball paradigms as assessed using fNIRS
Neurophotonics, 1(1). 015001
(26)
Neuropharmacological effect of methylphenidate on attention network in children with attention deficit/hyperactivity disorder during oddball paradigms as assessed using fNIRS
Neurophotonics, 1(1). 015001
Matsui, M., Homae, F., Tsuzuki, D., Watanabe, H., Katagiri, M., Uda, S., Nakashima, M., Dan, I. & Taga, G., (2014).
Referential framework for transcranial anatomical correspondence for fNIRS based on manually traced sulci and gyri of an infant brain
Neuroscience research, 80. 55-68
(25)
Referential framework for transcranial anatomical correspondence for fNIRS based on manually traced sulci and gyri of an infant brain
Neuroscience research, 80. 55-68
Tsuzuki, D.* & Dan, I., (2014).
Spatial registration for functional near-infrared spectroscopy: From channel position on the scalp to cortical location in individual and group analyses
NeuroImage, 85. 92-103
(24)
Spatial registration for functional near-infrared spectroscopy: From channel position on the scalp to cortical location in individual and group analyses
NeuroImage, 85. 92-103
Dan, H., Dan, I., Sano, T., Kyutoku, Y., Oguro, K., Yokota, H., Tsuzuki, D., & Watanabe, E., (2013).
Language-specific cortical activation patterns for verbal fluency tasks in Japanese as assessed by multichannel functional near-infrared spectroscopy
Brain and language, 126(2). 208-216
(23)
Language-specific cortical activation patterns for verbal fluency tasks in Japanese as assessed by multichannel functional near-infrared spectroscopy
Brain and language, 126(2). 208-216
Kimura, A., Wada, Y., Masuda, T., Goto, S-i., Tsuzuki, D., Hibino, H., Cai, D., & Dan, I., (2013).
Memory Color Effect Induced by Familiarity of Brand Logos
PLoS ONE, 8(7) e68474. doi:10.1371/journal.pone.0068474.
(22)
Memory Color Effect Induced by Familiarity of Brand Logos
PLoS ONE, 8(7) e68474. doi:10.1371/journal.pone.0068474.
Watanabe, H., Homae, F., Nakano, T., Tsuzuki, D., Enkhtur, L., Nemoto, K., Dan, I., & Taga, G., (2013).
Effect of auditory input on activations in infant diverse cortical regions during audiovisual processing
Human Brain Mapping, 34(3). 543-565
(21)
Effect of auditory input on activations in infant diverse cortical regions during audiovisual processing
Human Brain Mapping, 34(3). 543-565
Saito, T., Uga, M., Tsuzuki, D., Yokota, H., Oguro K., Yamamoto, T., Dan, I., & Watanabe E., (2013).
Evoked potential mapping of the rostral region by frameless navigation system in Mexican hairless pig
Journal of Neuroscience Methods, 212(1). 100-105
(20)
Evoked potential mapping of the rostral region by frameless navigation system in Mexican hairless pig
Journal of Neuroscience Methods, 212(1). 100-105
Monden, Y., Dan, H., Nagashima, M., Dan, I., Tsuzuki, D., Kyutoku, Y., Gunji, Y., Yamagata, T., Watanabe, E., & Momoi, M., (2012).
Right prefrontal activation as a neuro-functional biomarker for monitoring acute effects of methylphenidate in ADHD children: An fNIRS study
NeuroImage: CLINICAL, 1(1). 131-140
(19)
Right prefrontal activation as a neuro-functional biomarker for monitoring acute effects of methylphenidate in ADHD children: An fNIRS study
NeuroImage: CLINICAL, 1(1). 131-140
Otsuka, T., Dan, H., Dan, I., Sase, M., Sano, T., Tsuzuki, D., Fujita, A., Sasaguri, K., Okada, N., Kusama, M., Jinbu, Y., & Watanabe, E., (2012).
Effect of Local Anesthesia on Trigeminal Somatosensory-evoked Magnetic Fields
Journal of Dental Research, 91(12). 1196-1201
(18)
Effect of Local Anesthesia on Trigeminal Somatosensory-evoked Magnetic Fields
Journal of Dental Research, 91(12). 1196-1201
Cooper, RJ., Caffini, M., Dubb, J., Fang, Q., Custo, A., Tsuzuki, D., Fischl, B., Wells, W 3rd, Dan, I., & Boas, DA., (2012).
Validating atlas-guided DOT: A comparison of diffuse optical tomography informed by atlas and subject-specific anatomies
NeuroImage, 62(3). 1999-2006
(17)
Validating atlas-guided DOT: A comparison of diffuse optical tomography informed by atlas and subject-specific anatomies
NeuroImage, 62(3). 1999-2006
Moriai-Izawa, A., Dan, H., Dan, I., Sano, T., Oguro, K., Yokota, H., Tsuzuki, D., & Watanabe, E., (2012).
Multichannel fNIRS assessment of overt and covert confrontation naming
Brain and Language, 121(3). 185-193
(16)
Multichannel fNIRS assessment of overt and covert confrontation naming
Brain and Language, 121(3). 185-193
Mizutani, N., Dan, I., Kyutoku, Y., Tsuzuki, D., Clowney, L., Kusakabe, Y., Okamoto, M., & Yamanaka, T., (2012).
Package images modulate flavors in memory: Incidental learning of fruit juice flavors
Food Quality and Preference, 24. 92-98
(15)
Package images modulate flavors in memory: Incidental learning of fruit juice flavors
Food Quality and Preference, 24. 92-98
Tsuzuki, D., Cai, D., Dan, H., Kyutoku, Y., Fujita, A., Watanabe, E., & Dan, I., (2012).
Stable and convenient spatial registration of stand-alone NIRS data through anchor-based probabilistic registration
Neuroscience research, 72(2). 163-171
(14)
Stable and convenient spatial registration of stand-alone NIRS data through anchor-based probabilistic registration
Neuroscience research, 72(2). 163-171
Sugiura, L., Ojima, S., Matsuba-Kurita, H., Dan, I., Tsuzuki, D., Katura, T., & Hagiwara, H., (2011).
Sound to Language: Different Cortical Processing for First and Second Languages in Elementary School Children as Revealed by a Large-Scale Study Using fNIRS
Cerebral Cortex, 21(10). 2374-2393
(13)
Sound to Language: Different Cortical Processing for First and Second Languages in Elementary School Children as Revealed by a Large-Scale Study Using fNIRS
Cerebral Cortex, 21(10). 2374-2393
Nemoto, K., Dan, I., Rorden, C., Ohnishi, T., Tsuzuki, D., Okamoto, M., Yamashita, F., & Asada, T., (2011).
Lin4Neuro: a customized Linux distribution ready for neuroimaging analysis
BMC Medical Imaging, doi:10.1186/1471-2342-11-3. 2011
(12)
Lin4Neuro: a customized Linux distribution ready for neuroimaging analysis
BMC Medical Imaging, doi:10.1186/1471-2342-11-3. 2011
Kimura, A., Wada, Y., Kamada, A., Masuda, T., Okamoto, M., Goto, S., Tsuzuki, D., Cai, D., Oka, T., & Dan, I., (2010).
Interactive effects of carbon footprint information and its accessibility on value and subjective qualities of food products
Appetite, 55. 271-278
(11)
Interactive effects of carbon footprint information and its accessibility on value and subjective qualities of food products
Appetite, 55. 271-278
Kimura, A., Wada, Y., Ohshima, K., Yamaguchi, Y., Tsuzuki, D., Oka, T., & Dan, I., (2010).
Eating habits in childhood relate to preference for traditional diets among young Japanese
Food Quality and Preference, 843-848.
(10)
Eating habits in childhood relate to preference for traditional diets among young Japanese
Food Quality and Preference, 843-848.
Yanagisawa, H., Dan, I., Tsuzuki, D., Kato, M., Okamoto, M., Kyutoku, Y., & Soya, H., (2010).
Acute moderate exercise elicits increased dorsolateral prefrontal activation and improves cognitive performance with Stroop test
NeuroImage, 50. 1702-1710
(9)
Acute moderate exercise elicits increased dorsolateral prefrontal activation and improves cognitive performance with Stroop test
NeuroImage, 50. 1702-1710
Wada, Y., Arce-Lopera, C., Masuda, T., Kimura, A., Dan, I., Goto S., Tsuzuki, D., & Okajima, K., (2010).
Influence of luminance distribution on the appetizingly fresh appearance of cabbage
Appetite, 54. 363-368
(8)
Influence of luminance distribution on the appetizingly fresh appearance of cabbage
Appetite, 54. 363-368
Custo, A., Boas, DA., Tsuzuki, D., Dan, I., Mesquita, R., Fischl, B., Grimson, WE., & Wells, W 3rd., (2010).
Anatomical atlas-guided diffuse optical tomography of brain activation
NeuroImage, 49. 561-567
(7)
Anatomical atlas-guided diffuse optical tomography of brain activation
NeuroImage, 49. 561-567
Okamoto, M., Tsuzuki, D., Clowney, L., Dan, H., Singh, K., & Dan, I., (2009).
Structural atlas-based spatial registration for functional near-infrared spectroscopy enabling inter-study data integration
Clinical Neurophysiology, 120. 1320-1328
(6)
Structural atlas-based spatial registration for functional near-infrared spectroscopy enabling inter-study data integration
Clinical Neurophysiology, 120. 1320-1328
Kimura, A., Wada, Y., Goto, S., Tsuzuki, D., Cai, D., Oka, T., & Dan, I., (2009).
Implicit gender-based food stereotypes: semantic priming experiments on young Japanese
Appetite, 52. 521-524
(5)
Implicit gender-based food stereotypes: semantic priming experiments on young Japanese
Appetite, 52. 521-524
Kimura, A., Wada, Y., Tsuzuki, D., Goto, S., Cai, D., & Dan, I., (2008).
Consumer valuation of packaged foods. Interactive effects of amount and accessibility of information
Appetite, 51. 628-634
(4)
Consumer valuation of packaged foods. Interactive effects of amount and accessibility of information
Appetite, 51. 628-634
Kubota, M., Inouchi, M., Dan, I., Tsuzuki, D., Ishikawa, A., & Scovel, T., (2008).
Fast (100-175 ms) components elicited bilaterally by language production as measured by three-wavelength optical imaging
Brain Research, 1226. 124-133
(3)
Fast (100-175 ms) components elicited bilaterally by language production as measured by three-wavelength optical imaging
Brain Research, 1226. 124-133
Wada, Y., Tsuzuki, D., Kobayashi, N., Hayakawa, F., & Kohyama, K., (2007).
Visual illusion in mass estimation of cut food
Appetite, 49. 183-190
(2)
Visual illusion in mass estimation of cut food
Appetite, 49. 183-190
Tsuzuki, D.#, Jurcak, V.#, Singh, A. K., Okamoto, M., Watanabe, E., & Dan, I. (# Equal contribution), (2007).
Virtual spatial registration of stand-alone fNIRS data to MNI space
NeuroImage, 34. 1506-1518
(1)
Virtual spatial registration of stand-alone fNIRS data to MNI space
NeuroImage, 34. 1506-1518
Jurcak, V.#, Tsuzuki, D.#, & Dan, I. (# Equal contribution), (2007).
10/20, 10/10, and 10/5 systems revisited: their validity as relative head-surface-based positioning systems
NeuroImage, 34. 1600-1611
10/20, 10/10, and 10/5 systems revisited: their validity as relative head-surface-based positioning systems
NeuroImage, 34. 1600-1611
 [NFRI functions howto]
[NFRI functions howto] Basic preparation for using NFRI_functions
NFRI_functions are set of MATLAB functions to perform probabilistic registrations according to the paper by Singh AK, Okamoto M et al. Neuroimage 27, 842-851 (2005). Please start by downloading the zip file and ReadMe file indicated below.
First of all, please download and unzip the archive, "nfri_functions.zip". If it didn't go wrong, the "nfri_functions" folder will be created. You can move this folder to anywhere you would like. However, there is one thing you should be careful about. AVOID USING FOLDER NAME with MULTIBYTE CHARACTERS in the path to the folder so that Matlab can well find the appropriate folder.
You need Matlab to run the tools. Version 7.1 or later would work fine. Run Matlab, and go to the "nfri_functions" folder or set a path to it. Then, you are ready for enjoying the tools.
ReadMe file, "ReadMe091114.pdf", is also available here.
First of all, please download and unzip the archive, "nfri_functions.zip". If it didn't go wrong, the "nfri_functions" folder will be created. You can move this folder to anywhere you would like. However, there is one thing you should be careful about. AVOID USING FOLDER NAME with MULTIBYTE CHARACTERS in the path to the folder so that Matlab can well find the appropriate folder.
You need Matlab to run the tools. Version 7.1 or later would work fine. Run Matlab, and go to the "nfri_functions" folder or set a path to it. Then, you are ready for enjoying the tools.
ReadMe file, "ReadMe091114.pdf", is also available here.
Individual spatial analysis with "nfri_mni_estimation"
This function probabilistically registers individual subject's NIRS probe positions obtained in a real-world coordinates to the MNI space. Instead of using subject's own MRI dataset, which is not obtained in a typical NIRS measurement, the function pick up brains in the reference database, and "probabilistically" registers the subject's NIRS probe onto the MNI-152 compatible canonical brain that is optimized for NIRS analysis. For each NIRS probe position, a set of MNI coordinates (x, y, z) with an error estimate (standard deviation) will be calculated.
Further information is available in "ReadMe091114.pdf" file indicated above.
Further information is available in "ReadMe091114.pdf" file indicated above.
Group spatial analysis with"MultiSubject4_0"
This tool sums up the results of individual analyses to yield group analysis data. This is just a beta version, so stored in an independent directory. But it works fine.
Further information is available in "ReadMe091114.pdf" file indicated above.
Further information is available in "ReadMe091114.pdf" file indicated above.
Anatomical labeling with "anatomlabel_final"
This function reads a list of MNI coordinate values and estimates anatomical labeling using several anatomical labels available for academic use. Please be reminded that we just assembled them for easier access and you have to respect the original developers. So, cite the original articles when you use them. Appropriate citations are as follows:
1) AAL(automatic anatomical labeling): Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., Mazoyer, B., Joliot, M., 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 15 (1), 273-289.
2) Brodmann area (Chris rorden' MRIcro): Rorden, C., Brett, M., 2000. Stereotaxic display of brain lesions. Behav. Neurol. 12, 191-200.
3) LPBA40: Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 2007; 39: 1064-1080.
4) Brodmann area (Talairach daemon): Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT. Automated Talairach atlas labels for functional brain mapping. Human Brain Mapping 2000; 10: 120-131.
Further information is available in "ReadMe091114.pdf" file indicated above.
1) AAL(automatic anatomical labeling): Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., Mazoyer, B., Joliot, M., 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 15 (1), 273-289.
2) Brodmann area (Chris rorden' MRIcro): Rorden, C., Brett, M., 2000. Stereotaxic display of brain lesions. Behav. Neurol. 12, 191-200.
3) LPBA40: Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 2007; 39: 1064-1080.
4) Brodmann area (Talairach daemon): Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT. Automated Talairach atlas labels for functional brain mapping. Human Brain Mapping 2000; 10: 120-131.
Further information is available in "ReadMe091114.pdf" file indicated above.
Plotting functional data onto MNI space with "nfri_mni_plot" and "nfri_mni_plot_imp"
The function, nfri_mni_plot, is a handy plotter that primarily exhibits cortical activation data onto the MNI-compatible canonical brain. Activation data can be any values such as t-value, F-value, regression coefficient, mean amplitude and so on. They are expressed according to a color scale. The size of plotted channel or probe positions can also be varied. Missing data are exhibited in gray. Out-of-range data can also be expressed wither in black and white.
On the other hand, "nfri_mni_plot_imp" function is a slightly modified version of nfri_mni_plot, generating impressionistic drawing of data. The edge of a sphere is blended with background color to yield impressionism-painting-like outlook. It works best with uniform size spheres to be drawn, and usually serves for final data preparation for a conference presentation or journal article. The usage is totally the same as nfri_mni_plot. However, nfri_mni_plot is much faster, especially for a large number of channels.
Further information is available in "ReadMe091114.pdf" file indicated above.
On the other hand, "nfri_mni_plot_imp" function is a slightly modified version of nfri_mni_plot, generating impressionistic drawing of data. The edge of a sphere is blended with background color to yield impressionism-painting-like outlook. It works best with uniform size spheres to be drawn, and usually serves for final data preparation for a conference presentation or journal article. The usage is totally the same as nfri_mni_plot. However, nfri_mni_plot is much faster, especially for a large number of channels.
Further information is available in "ReadMe091114.pdf" file indicated above.
Tools for realizing spatial registration on POTATo (Platform for Optical Topography Analysis Tools)
This is a tentative release of the POTATo tools. More precise and user-friendly instruction will appear later.
Click here for download.
Click here for download.
MNI coordinates and Macro-anatomical labels for 10/20, 10/10 and 10/5 positions
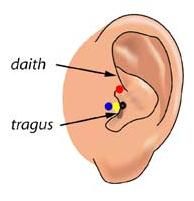
Here are lists of MNI coordinates and macroanatomical labels for 10/20, 10/10 and 10/5 positions. Please read the reference by Jurcack, Tsuzuki and Dan, NeuroImage (2007) for detailed information. You first need to choose either of the following pre-auricular points to define particular 10/20-based systems that you would like to adopt. Please check out the following figure.
Download the excel files as listed below for your reference.
1) Anterior root of the center of the peak region of tragus (=UI-10/5); Blue
2) Dent between upper edge of tragus and daith; Red
3) External ear canal; Black

Download the excel files as listed below for your reference.
1) Anterior root of the center of the peak region of tragus (=UI-10/5); Blue
2) Dent between upper edge of tragus and daith; Red
3) External ear canal; Black
 [Anchor-based probabilistic registration]
[Anchor-based probabilistic registration] Introduction
Anchor-based probabilistic registration is yet another spatial registration method for fNIRS study. It enables the normalization of cortical channel positions of subject onto MNI standardized space without one's own MRI. Initially, I implemented the anchor-based probabilistic registration software by Matlab. After publishing the paper, I have modified the script also to be run on Octave. If you don't have Matlab license, just feel free to install the latest version of Octave and try anchor-based probabilistic registration. You can download the archive here. Enjoy!
Usage.
[affinEstData, loadedData] = anchor_registration(Nz, AR, AL, Anchor, OtherP);
or, if you already have the data formatted in 'loadedData', just try this.
[affinEstData, loadedData] = anchor_registration(loadedData);
Input.
Nz ... <1x3> double, coordinate of Nasion
AR ... <1x3> double, coordinate of Right preauricular point
AL ... <1x3> double, coordinate of Left preauricular point
Anchor ... double, anchor point(s) around Cz on the head surface
OtherP ... double, coordinates of optodes or channels on the head surfacen
preauricular ... 'anterior_root_of_the_tragus' or 'dent' is available.
If this argument is ommited,
'anterior_root_of_the_tragus' is
automatically selected and it's coordinates are
applied for affine transformation.
loadedData.
D: [refpn x 3 double] ... [Nz; AR; AL; Anchor]
DDD: [nx3 double] ... OtherP
RW: [refpn x 3 x 17 double] ...
Anchor point for each head in NFRI_R17(RW)
N: [refpn x 3 x 17 double] ...
Anchor point for each head in NFRI_R17(MNI)
preauricular: Location of preauricular point.
'anterior_root_of_the_tragus' or 'dent' is available.
Output.
affinEstData.
OtherH: [nx3 double]
OtherC: [nx3 double]
OtherHSD: [nx4 double]
OtherCSD: [nx4 double]
SSwsH: [nx4 double]
SSwsC: [nx4 double]
RefN: 17
D: [refpn x 3 double]
DDD: [nx3 double]
N: [refpn x 3 x 17 double]
Example.
Nz = [ 0 -86.61 0];
AR = [-73.64 0 0];
AL = [ 73.72 0 0];
Anchor = [-4.1 -34.3 143.41];
OtherP = [49.16 -68.07 95.92
66.28 -42.55 100.08
75.29 -13.25 104.16];
[affinEstData, loadedData] = anchor_registration(Nz, AR, AL, Anchor, OtherP);
 [Collaborators and mentors]
[Collaborators and mentors] With many thanks!
Asakawa, Shin-ichi  | Bhat, Anjana
| Bhat, Anjana 
 | Boas, David
| Boas, David  | Cooper, Robert
| Cooper, Robert  | Custo, Anna
| Custo, Anna 
 | Dan, Ippeita
| Dan, Ippeita  | Dubb, Jay
| Dubb, Jay  | Fang, Qianqian
| Fang, Qianqian  | Gima, Hirotaka | Hagiwara, Hiroko (Died: July 10, 2015)
| Gima, Hirotaka | Hagiwara, Hiroko (Died: July 10, 2015)  | Homae, Fumitaka | Jurcak, Valer | Kajiyama, Yûji
| Homae, Fumitaka | Jurcak, Valer | Kajiyama, Yûji  | Katura, Takushige
| Katura, Takushige  | Kimura, Atsushi | Kyutoku, Yasushi | Masuda, Tomohiro | Matsui, Mie
| Kimura, Atsushi | Kyutoku, Yasushi | Masuda, Tomohiro | Matsui, Mie 
 | Meryem Yücel
| Meryem Yücel  | Monden, Yukifumi
| Monden, Yukifumi  | Minagawa, Yasuyo
| Minagawa, Yasuyo  | Nagashima, Masako
| Nagashima, Masako  | Nemoto, Kiyotaka
| Nemoto, Kiyotaka 
 | Noguchi, Kaoru (Died: Jul 25, 2006 at the age of 71) | Okamoto, Masako
| Noguchi, Kaoru (Died: Jul 25, 2006 at the age of 71) | Okamoto, Masako  | Singh, Archana
| Singh, Archana  | Sugiura, Lisa
| Sugiura, Lisa  | Takahashi, Nozomi
| Takahashi, Nozomi  | Taga, Gentaro | Uga, Minako | Wada, Yûji
| Taga, Gentaro | Uga, Minako | Wada, Yûji  | Watanabe, Eiju
| Watanabe, Eiju  | Watanabe, Hama | Yamada, Hiroshi (Died: May 28, 2015 at the age of 57)
| Watanabe, Hama | Yamada, Hiroshi (Died: May 28, 2015 at the age of 57)
 | Bhat, Anjana
| Bhat, Anjana 
 | Boas, David
| Boas, David  | Cooper, Robert
| Cooper, Robert  | Custo, Anna
| Custo, Anna 
 | Dan, Ippeita
| Dan, Ippeita  | Dubb, Jay
| Dubb, Jay  | Fang, Qianqian
| Fang, Qianqian  | Gima, Hirotaka | Hagiwara, Hiroko (Died: July 10, 2015)
| Gima, Hirotaka | Hagiwara, Hiroko (Died: July 10, 2015)  | Homae, Fumitaka | Jurcak, Valer | Kajiyama, Yûji
| Homae, Fumitaka | Jurcak, Valer | Kajiyama, Yûji  | Katura, Takushige
| Katura, Takushige  | Kimura, Atsushi | Kyutoku, Yasushi | Masuda, Tomohiro | Matsui, Mie
| Kimura, Atsushi | Kyutoku, Yasushi | Masuda, Tomohiro | Matsui, Mie 
 | Meryem Yücel
| Meryem Yücel  | Monden, Yukifumi
| Monden, Yukifumi  | Minagawa, Yasuyo
| Minagawa, Yasuyo  | Nagashima, Masako
| Nagashima, Masako  | Nemoto, Kiyotaka
| Nemoto, Kiyotaka 
 | Noguchi, Kaoru (Died: Jul 25, 2006 at the age of 71) | Okamoto, Masako
| Noguchi, Kaoru (Died: Jul 25, 2006 at the age of 71) | Okamoto, Masako  | Singh, Archana
| Singh, Archana  | Sugiura, Lisa
| Sugiura, Lisa  | Takahashi, Nozomi
| Takahashi, Nozomi  | Taga, Gentaro | Uga, Minako | Wada, Yûji
| Taga, Gentaro | Uga, Minako | Wada, Yûji  | Watanabe, Eiju
| Watanabe, Eiju  | Watanabe, Hama | Yamada, Hiroshi (Died: May 28, 2015 at the age of 57)
| Watanabe, Hama | Yamada, Hiroshi (Died: May 28, 2015 at the age of 57)
Undergraduate students (2023)
TAKAHARA, Sana 
食器と食品の彩度対比が味覚イメージに及ぼす影響
植物の病気の診断における機械学習の応用
静止画を対象とした CNN による車両認識
胸部 X 線画像を対象とした機械学習による病変分類の性能評価 - 実行環境が及ぼす影響の比較検討 -

食器と食品の彩度対比が味覚イメージに及ぼす影響
植物の病気の診断における機械学習の応用
静止画を対象とした CNN による車両認識
胸部 X 線画像を対象とした機械学習による病変分類の性能評価 - 実行環境が及ぼす影響の比較検討 -
Undergraduate students (2016)
HOUHASHI, Manami 
機械学習による相貌認知へのアプローチ〜顔画像を対象としたディープラーニングによるキレイ・カワイイの判断〜
8bit 家庭用ゲーム機をモチーフとした顔画像抽象化システムの開発
いまだ「左の顔」優位の傾向は変わっていないのか?~情報工学科 5 年生の美的感覚に迫る~

機械学習による相貌認知へのアプローチ〜顔画像を対象としたディープラーニングによるキレイ・カワイイの判断〜
8bit 家庭用ゲーム機をモチーフとした顔画像抽象化システムの開発
いまだ「左の顔」優位の傾向は変わっていないのか?~情報工学科 5 年生の美的感覚に迫る~